Some backgroundPermalink
Lucky you! You just got hold of a largish CSV file (let’s say 15 GB, about 140 million rows). How do you handle this file to be able to work with it using Apache Arrow?
Going through the documentation of Arrow, you might notice that several ways are mentioned to import data. They fall into two families:
- one that I will refer to as the Single file API1;
- the other is the Dataset API.
The Single file API contains functions for each supported file format (CSV, JSON, Parquet, Feather/Arrow, ORC). They work on one file at a time, and they load the data in memory. So depending on the size of your file and the amount of memory you have available on your system, it might not be possible to load the dataset this way. If you can load the dataset in memory queries will run faster because the data will be readily accessible to the query engine.
The Dataset API is very flexible. It can read multiple file formats, you can point to a folder with multiple files and create a dataset from them, and it can read datasets from multiple sources (even combining remote and local sources). This API can also be used to read single files that are too large to fit in memory. This works because the files are not actually loaded in memory. The functions scan the content so they know where to look for the data and what the schema is (the data types and names of each column). When you query the data, there is some overhead because the query engine needs to first read the data before it can operate on it. (If you want to see some examples of what the Dataset API can do, check out the two previous posts on datasets with Arrow: Part 1, and Part 2)
In this post, we will explore how to convert a large CSV file to the Apache Parquet format using the Single file and the Dataset APIs with code examples in R and Python. We do the conversion from CSV to Parquet, because in a previous post we found that the Parquet format provided the best compromise between disk space usage and query performance. Having the content of this file in the Apache Parquet format will ensure that we can read and operate on this data quickly.
The Single file API in RPermalink
The functions in the Single file API in R start with read_ or
write_ followed by the name of the file format. For instance,
read_csv_arrow(), read_parquet(), and read_feather() belong to
what I refer here as the Single file API.
To read the data with our 15 GB CSV file, we would use:
library(arrow)
data <- read_csv_arrow(
"~/dataset/path_to_file.csv",
as_data_frame = FALSE
)
Using as_data_frame = FALSE keeps the result as an Arrow table which
is a better representation for a file of this size. Attempting to
convert it into a data frame will take longer to load, and you will
most likely run out of memory.
This step takes about 15 seconds on my system. As far as I can tell, the arrow R package is the only way to load a file of this size in memory. Both readr/vroom and data.table ran out of memory after several minutes and before being able to finish reading the file.
At this point, you have an Arrow formatted table loaded in memory that is ready for you to work with.
To convert this file into the Apache Parquet format using the Single file API, you would use:
write_parquet(data, "~/dataset/data.parquet")
Creating this file takes about 85 seconds on my system. The resulting file is about 9.5 GB, reducing the amount of hard drive space needed to store the data by about 60%.
The read_parquet() function will load this dataset the next time you
need to work with it:
data <- read_parquet("~/dataset/data.parquet", as_data_frame = FALSE)
Let’s count the number of unique values in one of the columns of this dataset:
data %>%
count(variable) %>%
collect()
This query takes only half a second on my laptop. Half a second to summarize the content of 140 million rows: this is fast! Very fast!
Whether you use read_csv_arrow() or read_parquet(), the dataset is
loaded in memory using the same representation: an Arrow table. The
performance of queries would therefore be the same regardless of the
format used to store the data. In this case, the decision to storing
the data as a CSV or a Parquet file will be based on the amount of
storage, how fast reading from CSV or Parquet compares to the overhead
associated with the conversion from one format to the other.
Let’s now use the Dataset API.
The Dataset API in RPermalink
We will read the large CSV file with open_dataset(). This function
can be pointed to a folder with several files but it can also be used
to read a single file.
data <- open_dataset("~/dataset/path_to_file.csv")
With our 15 GB file, it takes 0.05 seconds to “read” the file. It is
fast because the data does not get loaded in memory. open_dataset()
scans the content of the file to identify the name of the columns and
their data types.
Running the same query as above, which counts the number of unique values in a column, takes 18 seconds compared to the 0.5 seconds when the data is loaded in memory. It is slower because the query engine needs to read the data. It is the same result that we had found in a previous post: running queries directly on a CSV file is slow. In that post, we also found that storing the data in the Parquet format sped things up. Let’s now convert this dataset to Parquet using the Dataset API.
Instead of using a single Parquet file as we did above when we looked
at the Single file API, we will also partition the Parquet dataset to
see how it could help with query performance. The particular dataset I
have on hand does not have any obvious variable we can use to
partition the data. If you are dealing with a dataset that has
timestamps for data collected at regular intervals, partitioning on a
temporal dimension could make sense (that’s what the NYC taxi dataset
does by partitioning by year and month). Instead, here, we can use the
max_rows_per_file argument of the write_dataset() function to
limit how large each Parquet file is. At least for this dataset, I
found that limiting the number of rows to 10 million per file seemed
like a good compromise. Each file is about 720 MB which is close to
the file sizes in the NYC taxi dataset. The PyArrow
documentation
has a good overview of strategies for partitioning a dataset. The
general recommendation is to avoid individual Parquet files smaller
than 20 MB and larger than 2 GB while avoiding a partition layout that
would create more than 10,000 partitions.
write_dataset(
data,
format = "parquet",
path = "~/datasets/my-data/",
max_rows_per_file = 1e7
)
Writing these files on my system takes about 50 seconds. We end up with 14 Parquet files totaling 9.9 GB.
Next time we want to work with this data, we can load these files with:
data <- open_dataset(
"~/datasets/my-data"
)
It takes about the same amount of time as scanning the CSV files. It is almost instantaneous taking only 0.02 seconds. Again, this is fast because the data is not loaded in memory. We saw that with this approach it took almost 20 seconds to run this query on our CSV file. So what is the performance of a query on this dataset split into multiple Parquet files?
Counting the unique values in a column takes just 1 second. You read that correctly. One second to summarize 140 million rows. It is a little slower than doing it when the entire dataset is loaded in memory but scanning the files is faster. And because the dataset is not loaded in memory, you are not limited by the amount of memory you have available. With the Single File API, a file of 15 GB is the upper limit of what my laptop with 32 GB of RAM can handle.
One of the advantages of the Arrow ecosystem is that it is polyglot. The approach we described with R also works with Python. And because both languages use the same C++ backend, the code looks very similar.
Single file API in PythonPermalink
There are two functions in the PyArrow Single API to read CSV files:
read_csv() and open_csv(). While read_csv() loads all the data
in memory and does it fast by using multiple threads to read different
parts of the files, open_csv() reads the data in batches and uses a
single thread.
If the CSV file is small enough, you should use read_csv(). The code
to read the CSV file and write it to a Parquet file would then look
like this:
import pyarrow as pa
import pyarrow.csv
import pyarrow.parquet as pq
in_path = '~/datasets/data.csv'
out_path = '~/datasets/data.parquet'
data = pa.csv.read_csv(in_path)
pq.write_table(data, out_path)
In our case, the file is too large to fit in memory2. So instead of
using read_csv(), we need to use open_csv(). Because, the CSV file
is read in chunks, the code is a little more complex. We need to loop
through each chunk, read it, and write it to the Parquet file. This
uses little memory but is not as fast as using read_csv(). While
read_csv() is multi-threaded, open_csv() uses a single
thread. When using open_csv(), the data types need to be consistent
in your columns. The function infers the data types on the first chunk
of data read, and if the type changes halfway through your dataset in
one of your columns, you will run into errors. You can avoid this by
specifying the data types manually.
# from <https://stackoverflow.com/a/68563617/1113276>
import pyarrow as pa
import pyarrow.parquet as pq
import pyarrow.csv
in_path = '~/datasets/data.csv'
out_path = '~/datasets/data.parquet'
writer = None
with pyarrow.csv.open_csv(in_path) as reader:
for next_chunk in reader:
if next_chunk is None:
break
if writer is None:
writer = pq.ParquetWriter(out_path, next_chunk.schema)
next_table = pa.Table.from_batches([next_chunk])
writer.write_table(next_table)
writer.close()
On my system, the conversion from file to Parquet takes about 190 seconds. Reading the Parquet file can be done with:
data = pq.ParquetDataset(out_path).read()
With this approach, the dataset is in memory, just like when we were using R. Again with 32 GB of RAM in my laptop, I need to be careful with what is running on my system to be able to load this dataset without running out of memory and crashing my Python session.
The Dataset API in PythonPermalink
To load the CSV file with the Dataset API, we use the dataset()
function:
import pyarrow.dataset as ds
in_path = "~/datasets/data.csv"
out_path = "~/datasets/my-data/"
data = ds.dataset(in_path)
Just like with R, importing this file takes about 0.02 seconds.
To convert it to a collection of Parquet files, you use the
write_dataset() function. This function takes the same
max_rows_per_file argument to control the size of the Parquet file
in each partition.
ds.write_dataset(data, out_path, format = "parquet",
max_rows_per_file = 1e7)
Reading this collection of Parquet files can also be done with the
dataset() function, just like when we used the function to read the
single CSV file above. The dataset() function is very flexible and
can be used to import data in a variety of formats, and structures,
and even combines files from local and remote locations. The format
argument is optional as the function detects automatically the file
type.
data = ds.dataset(out_path, format = "parquet")
Given the current functionalities implemented PyArrow, querying datasets of this size is possible but it is neither blazing fast nor convenient. A good alternative is to use Ibis with DuckDB as a backend. Ibis provides a single interface to work with data stored in memory or in databases. DuckDB is a self-contained database designed for data analytics. These tools deserve a lot more than a one-sentence summary but this is beyond the scope of this post.
To count the number of unique values, you could use the following approach:
import ibis
ibis.options.interactive = True
con = ibis.duckdb.connect()
data = con.register("parquet:///home/user/datasets/my-data/*.parquet", table_name = "table")
con.table("table").variable.value_counts()
Just like with using R, this takes about a second to count the unique values in one column of our 140 million row dataset.
What this post didn’t mentionPermalink
I focused on the reading of a CSV file and its conversion to Parquet. I didn’t talk about all the options that both the Single file and the Dataset APIs have to customize the format of the files that are being imported. For instance, both APIs can be used to specify a different column-separator, and cell content that should be treated as missing data.
ConclusionPermalink
For a 15 GB data file, the dataset API is better suited to read, convert, and query the data. There is an overhead associated with not having the data in memory but it is greatly reduced if the data is stored as Parquet files. Another advantage is that the approach developed here would scale to much larger datasets where the Single file API would not be able to serialize the data in memory.
With the dataset in this example, the Single file API did not have an opportunity to shine given the hardware constraints of a modern laptop. However, if you are dealing with datasets that fit easily in memory, working with data directly in memory will lead to better query performance.
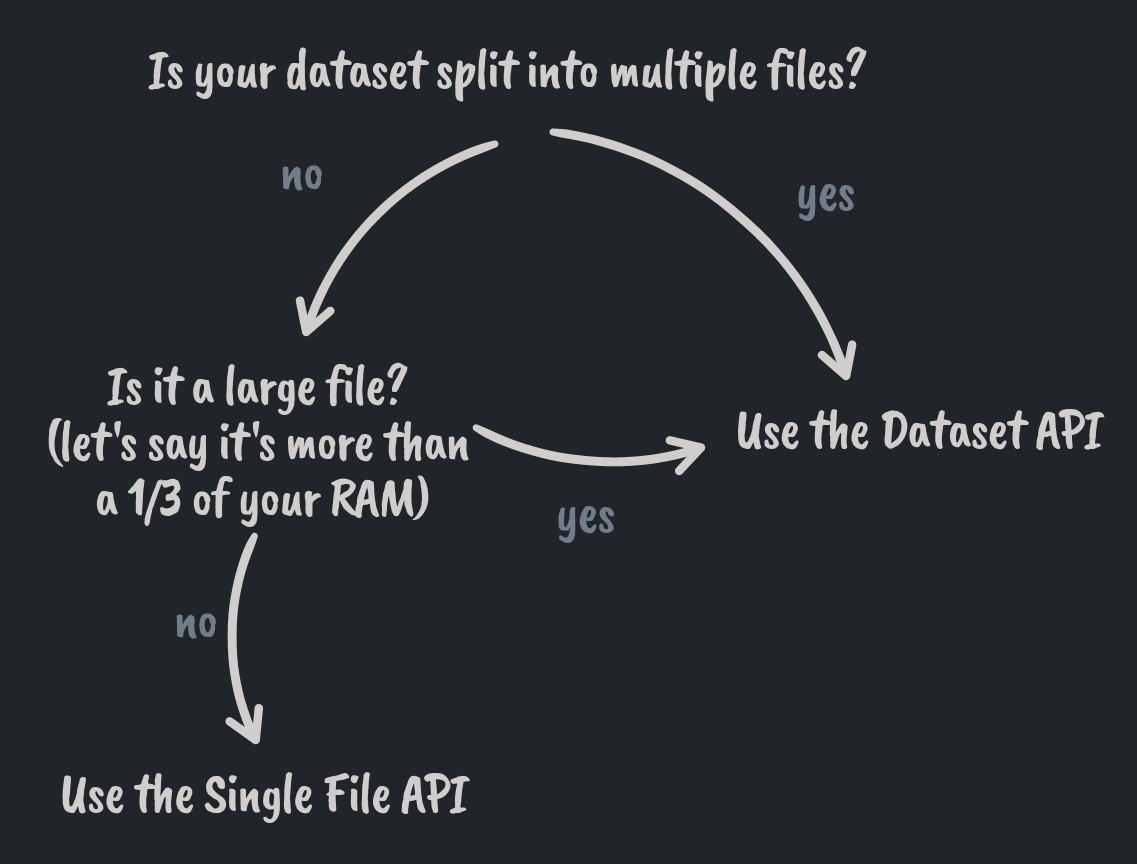
To summarize what we learned in this post, this brief decision guide to help you choose the correct API to import your data.
AcknowledgmentsPermalink
Thank you to Kae Suarez and Danielle Navarro for reviewing this post and providing feedback that improved its content.

Comments